Our research in this area has been funded by the European Research Council and the Synergy-Excellence programme of Sachsen-Anhalt with EFRE support. This is greatly appreciated.
Modeling, simulation, and optimization are important methodological pillars of computational science and have triggered a revolution in the way we approach things. Spearheaded by applications in mechanical engineering, logistics, chemical engineering, and information technologogy, the last two decades saw an explosion of computational approaches being applied to biology and medicine. On the one hand, they pose a bunch of additional challenges, such as the nonavailability of mathematical models, uncertainties, and often low quality measurement data. On the other hand there is an obvious large potential for more efficiency and a huge amount of available patient-specific data. This trend is further accelerated by availability of data, open source software, hardware improvements, and interdisciplinary approaches and cooperations. Our main contributions are on the level of clinical decision support. Using available data and clinical expert knowledge, we have been modeling relationships between different inputs and relevant biomarkers, and making them accessible for optimization approaches.
A survey paper on the interplay between clinical decision support and optimization was published in Optima. We recommend this paper for an overview of our models, methods, and case studies.
Clinical Decision Support for Blood Cancer
Our long term goal is to provide optimization-driven individual decision support for dosage and treatment scheduling, taking combinations of treatments and practical restrictions into account.
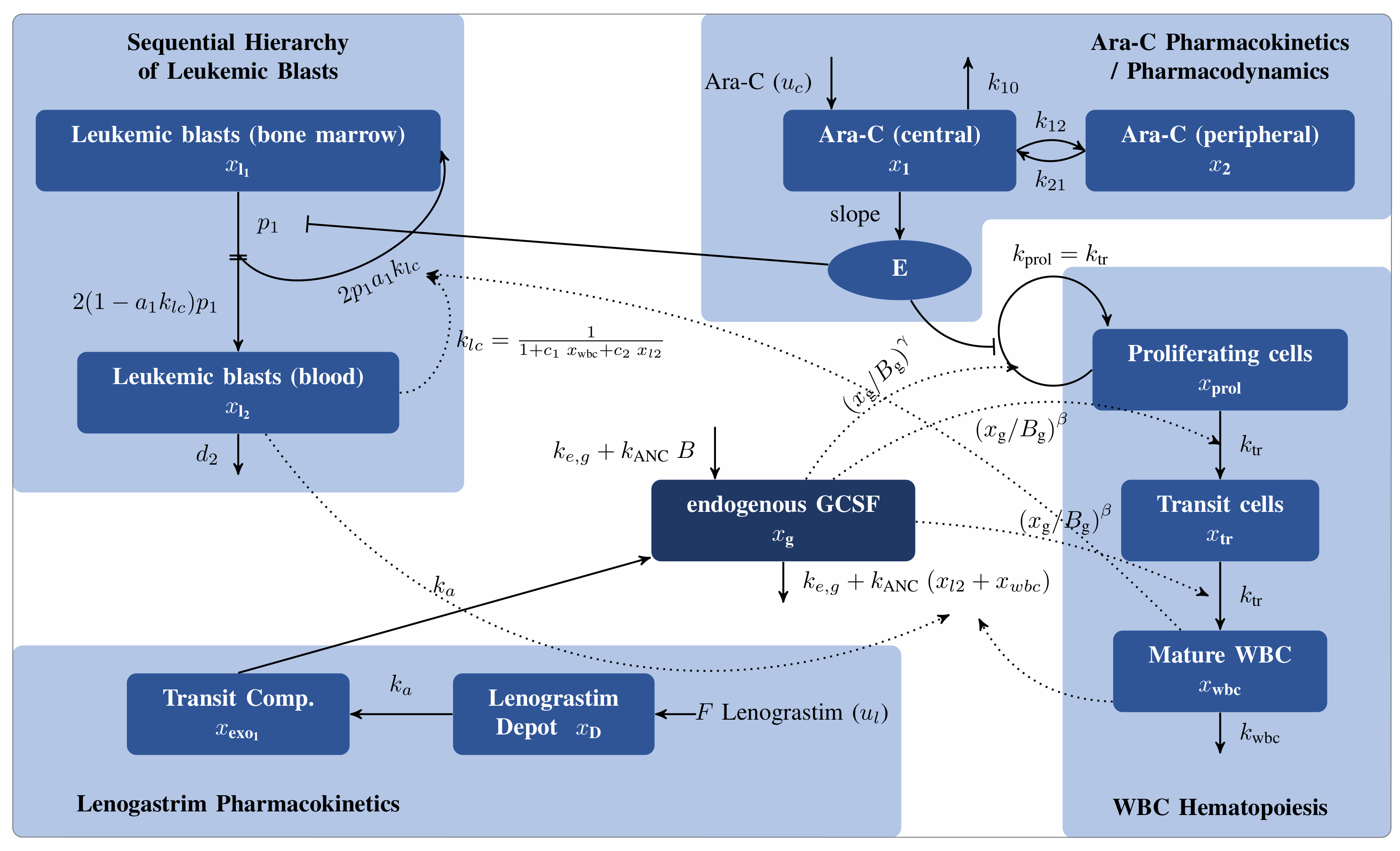
For example, we have been modeling the interplay between chemotherapy, healthy cells, immune system, and blasts in acute myeloid leukemia, as visualized in the following figure.
Mathematical model for AML treatment from Jost2021.
The differential equation model was developed, analyzed using advanced methods of parameter estimation, experimental design, uncertainty quantification, sensitivity analysis, and used to calculate personalized and mathematically optimal treatment plans in Jost et al., Model-based optimal AML consolidation treatment, 2021.
As a main result, the new approach can (in-silico) avoid 90% of occuring leukopenias without impairing the long term effect on the cancerous cells. The model makes obviously various simplifying assumptions. We work with different kinds of models for different purposes, following the digital twin paradigm. While above model showed to be a good compromise for real-world application and decision support, we have also developed more involved models based on partial differential equations, which are better suited to understand intrinsic and induced oscillations in the system. As a third line of research we consider machine learning models, with the ultimate goal to achieve accurate, explainable, and applicable models for clinical decision support. The same paradigm applies to other ongoing projects in oncology/hematology, e.g., scheduling of phlebotomia in polycythemia vera or chemotherapy treatment in pediatric acute lymphoblastic leukemia.
Discrimination between Atrial Fibrillation and Atrial Flutter
A similar approach of interacting models applies to applications in cardiology. Here we have been investigating different approaches to classify and treat cardiac arrhytmia. We have been developing and analyzing involved differential equations based on the Hodgkin-Huxley equations with the goal to understand and benchmark simplified mathematical models that can be used in hybrid machine learning approaches and for clinical decision support. As a byproduct of a new automatic classification approach to distinguish ventricular tachycardias, our new mathematical model presented in Sager et al. generalizes several classical and advanced block types (typical Type I block, atypical Type I block, the special cases of 2:1 and 3:2 Type I blocks, Type II block, advanced second-degree AV Block, and multi-level AV block.
We use mathematical optimization to discriminate atrial fibrillation from atypical atrial flutter, based on ECG data. We have been developing phenomenological mathematical models for multi-level blocks of signals in the AV node and optimization problems on top of them. The optimal objective function value gives an indication on whether an atypical atrial flutter may be the cause for the irregular signal in the ventricular chambers. We have also been developing tailored optimization algorithms to solve the arising mixed-integer nonlinear optimization problems efficiently.
Prediction of Earliest Activation of Focal Cardiac Arrhythmias
We investigate premature beats which are a common finding in patients suffering from structural heart disease, but they can also be present in healthy individuals. Catheter ablation represents a suitable therapeutic approach. However, the exact localization of the origin can be challenging, especially in cases of low PB burden during the procedure.
Our new algorithm is based on iterative regression analyses. When acquiring local activation times (LATs) within a 3-dimensional anatomic map of the corresponding heart chamber, this algorithm is able to identify that position where a next LAT measurement adds maximum information about the predicted site of origin. Furthermore, on the basis of the acquired LAT measurements, the algorithm is able to predict earliest activation with high accuracy.
A systematic retrospective analysis of the mapping performance comparing the operator with simulated search processes by the algorithm within 17 electroanatomic maps of focal spreading arrhythmias revealed a highly significant reduction of necessary LAT measurements from 55 to 10. Results can be found in this Heart Rhythm publication.
Decision support in Cardiac Resynchronization Therapy
The modern multimodal therapy of chronic heart disease with optimization of medication and device therapy improves the prognosis and quality of life. Pathophysiologically, cardiac decompensation is mediated and accompanied by severe neurohumoral dysregulations with consecutive further cardiac and renal irreversible damage. A decisive goal is the prevention of imminent decompensation or to mitigate its manifestation. For this purpose, it is necessary to specify predictors for the complex clinical and hemodynamic relationships with the goal of individualized therapy in the case of cardiac failure. The combination of disease-specific measurements and dynamic modeling approaches can improve sensitivity to characterize the regulatory system behavior. To model and simulate this pathology we want to extend a hemodynamic model with specific clinical measurements. The CircAdapt model describes the heart and circulation in a phenomenological way, calculating beat-to-beat hemodynamics and cardiac mechanics as well as focussing on the interactions of the ventricles via the septum. Thereby, several variables such as compliance and pulmonary vascular resistance should be mirrored. Especially, the longterm objective of this project is to improve the device based cardiac heart failure therapies.
Selected publications
@article{Reimann2026,
author = {Reimann, Adrian-Manuel and Schalk, Enrico and Mougiakakos, Dimitrios and Fischer, Thomas and Sager, Sebastian},
title = {Exploring innovative G-CSF schedules in AML cytarabine-based consolidation through a digital twin study of white blood cell recovery},
journal = {PLoS One},
publisher = {Springer},
year = {2026},
url = {https://www.medrxiv.org/content/10.1101/2025.07.17.25331695v1}
}
@article{Banga2024,
author = {Banga, Julio R and Sager, Sebastian},
title = {Generalized Inverse Optimal Control and its Application in Biology},
journal = {Annual Reviews of Control},
year = {2025},
url = {http://arxiv.org/abs/2405.20747},
doi = {10.1016/j.arcontrol.2025.101029}
}
@article{Gebhard2023,
author = {Gebhard, Anna and Lilienthal, Patrick and Metzler, Markus and Rauh, Manfred and Sager, Sebastian and Schmiegelow, Kjeld and Toksvang, Linea Natalie and Zierk, Jakob},
title = {Pharmacokinetic--pharmacodynamic modeling of maintenance therapy for childhood acute lymphoblastic leukemia},
journal = {Scientific Reports},
publisher = {Nature Publishing Group UK London},
year = {2023},
volume = {13},
number = {1},
pages = {11749},
url = {https://www.nature.com/articles/s41598-023-38414-0}
}
@article{Reimann2023,
author = {Reimann, Adrian-Manuel and Schalk, Enrico and Jost, Felix and Mougiakakos, Dimitrios and Weber, Daniela and D{\"o}hner, Hartmut and R{\'e}cher, Christian and Dumas, Pierre-Yves and Ditzhaus, Marc and Fischer, Thomas and Sager, Sebastian},
title = {AML consolidation therapy: timing matters},
journal = {Journal of Cancer Research and Clinical Oncology},
publisher = {Springer},
year = {2023},
volume = {149},
number = {15},
pages = {13811--13821},
url = {https://link.springer.com/article/10.1007/s00432-023-05115-0}
}
@article{Sager2023,
author = {Sager, Sebastian},
title = {Digital Twins in Oncology},
journal = {Journal of Cancer Research and Clinical Oncology},
publisher = {Springer},
year = {2023},
volume = {149},
number = {9},
pages = {5475--5477},
url = {https://link.springer.com/article/10.1007/s00432-023-05115-0}
}
@article{Minakowska2021,
author = {Minakowska, M. and Richter, T. and Sager, S.},
title = {A finite element / neural network framework for modeling suspensions of non-spherical particles - Concepts and medical applications},
journal = {Vietnam Journal of Mathematics},
year = {2021},
volume = {49},
pages = {207--235},
url = {https://arxiv.org/abs/2009.10818}
}
@article{Sager2021,
author = {Sager, Sebastian and Bernhardt, Felix and Kehrle, Florian and Merkert, Maximilian and Potschka, Andreas and Meder, Benjamin and Katus, Hugo and Scholz, Eberhard},
title = {Expert-Enhanced Machine Learning for Cardiac Arrhythmia Classification},
journal = {PloS one},
year = {2021},
volume = {16},
number = {12},
pages = {e0261571},
url = {https://optimization-online.org/?p=16046}
}
@article{Jost2020,
author = {Jost, Felix and Zierk, Jakob and Le, Thuy T.T. and Raupach, Thomas and Zierk, Jakob and Rauh, Manfred and Suttorp, Meinolf and Stanulla, Martin and Metzler, Markus and Sager, Sebastian},
title = {Model-based simulation of maintenance therapy of childhood acute lymphoblastic leukemia},
journal = {Frontiers in Physiology},
publisher = {Frontiers},
year = {2020},
volume = {11},
pages = {217},
url = {https://www.frontiersin.org/article/10.3389/fphys.2020.00217},
doi = {10.3389/fphys.2020.00217}
}
@article{Jost2020a,
author = {Jost, F. and Schalk, E. and Weber, D. and D\"ohner, H. and Fischer, T. and Sager, S.},
title = {Model-based optimal AML consolidation treatment},
journal = {IEEE Transactions on Biomedical Engineering},
year = {2020},
volume = {67},
number = {12},
pages = {3296--3306},
url = {https://arxiv.org/abs/1911.08980}
}
@article{Lilienthal2020,
author = {Lilienthal, P. and Tetschke, M. and Schalk, E. and Fischer, T. and Sager, S.},
title = {Optimized and Personalized Phlebotomy Schedules for Patients suffering from Polycythemia Vera},
journal = {Frontiers in Physiology},
publisher = {Frontiers},
year = {2020},
volume = {11},
pages = {328},
url = {http://journal.frontiersin.org/article/10.3389/fphys.2020.00328}
}
@article{Scholz2020,
author = {Scholz, E. and Hartlage, C. and Bernhardt, F. and Weber, T. and Salatzki, J. and Andr\'e, F. and Lugenbiel, P. and Riffel, J. and Katus, H. and Sager, S.},
title = {Spatial relationship between ablation sites within the pulmonary trunk and the left coronaries: Systematic risk assessment based on automated three-dimensional distance measurements},
journal = {Heart Rhytm O2},
year = {2020},
volume = {1.1},
pages = {14--20}
}
@article{Jost2019,
author = {Jost, F. and Schalk, E. and Rinke, K. and Fischer, T. and Sager, S.},
title = {Mathematical models for cytarabine-derived myelosuppression in acute myeloid leukaemia},
journal = {PLoS One},
publisher = {Public Library of Science},
year = {2019},
volume = {14},
number = {7},
pages = {e0204540},
doi = {10.1371/journal.pone.0204540}
}
@article{Sager2018,
author = {Sager, S.},
title = {Optimization and Clinical Decision Support},
journal = {Optima},
year = {2018},
volume = {104},
series = {Optima},
pages = {1--8},
organization = {Mathematical Optimisation Society},
url = {http://www.mathopt.org/Optima-Issues/optima104.pdf}
}
@article{Sager2018a,
author = {Sager, Sebastian},
title = {Optimierung und Klinische Entscheidungsunterst\"utzung},
journal = {Mitteilungen der Deutschen Mathematiker-Vereinigung},
year = {2018},
volume = {26},
pages = {101--111},
organization = {DMV},
doi = {10.1515/dmvm-2018-0030}
}
@article{Tetschke2018,
author = {Tetschke, M. and Lilienthal, P. and Pottgiesser, T. and Fischer, T. and Schalk, E. and Sager, S.},
title = {Mathematical Modeling of {RBC} Count Dynamics after Blood Loss},
journal = {Processes},
year = {2018},
volume = {6},
number = {9},
pages = {157--185},
doi = {10.3390/pr6090157}
}
@article{Weber2017,
author = {Weber, T. and Katus, H.A. and Sager, S. and Scholz, E.P.},
title = {{N}ovel algorithm for accelerated electroanatomical mapping and prediction of earliest activation of focal cardiac arrhythmias using mathematical optimization},
journal = {Heart Rhythm},
year = {2017},
volume = {14},
number = {6},
pages = {875--882},
url = {https://mathopt.de/publications/Weber2017.pdf},
doi = {10.1016/j.hrthm.2017.03.001}
}
@inproceedings{Zeile2017,
author = {Zeile, C. and Rauwolf, T. and Schmeisser, A. and Weber, T. and Sager, S.},
title = {The Influence of Right Ventricular Afterload in Cardiac Resynchronization Therapy: A CircAdapt Study},
journal = {Computing in Cardiology 2017 -PapersOnLine Proceedings},
year = {2017},
url = {http://www.cinc.org/archives/2017/pdf/215-232.pdf}
}
@article{Jost2016,
author = {Jost, Felix and Rinke, Kristine and Fischer, Thomas and Schalk, Enrico and Sager, Sebastian},
title = {Optimum Experimental Design for Patient Specific Mathematical Leukopenia Models},
journal = {IFAC-PapersOnLine},
publisher = {Elsevier},
year = {2016},
volume = {49},
number = {26},
pages = {344--349},
organization = {Magdeburg, Germany}
}
@article{Rinke2016,
author = {Rinke, Kristine and Jost, Felix and Findeisen, Rolf and Fischer, Thomas and Bartsch, Rainer and Schalk, Enrico and Sager, Sebastian},
title = {Parameter estimation for leukocyte dynamics after chemotherapy},
journal = {IFAC-PapersOnLine},
publisher = {Elsevier},
year = {2016},
volume = {49},
number = {26},
pages = {44--49},
organization = {Magdeburg, Germany}
}
@patent{Scholz2016,
author = {Scholz, E. and Sager, S. and Katus, H.},
title = {A system and computer program product for automatically distinguishing atrial flutter from atrial fibrillation},
year = {2016},
number = {EP2757940B1},
url = {https://patents.google.com/patent/EP2757940B1}
}
@article{Zeile2016,
author = {Zeile, Clemens and Scholz, Eberhard and Sager, Sebastian},
title = {A Simplified 2D Heart Model of the Wolff-Parkinson-White Syndrome},
journal = {IFAC-PapersOnLine},
publisher = {Elsevier},
year = {2016},
volume = {49},
number = {26},
pages = {26--31},
organization = {Magdeburg, Germany},
doi = {10.1016/j.ifacol.2016.12.098}
}
@article{Scholz2014,
author = {Scholz, E.P. and Kehrle, F. and Vossel, S. and Hess, A. and Zitron, E. and Katus, H.A. and Sager, S.},
title = {{D}iscriminating atrial flutter from atrial fibrillation using a multilevel model of atrioventricular conduction},
journal = {{H}eart {R}hythm},
year = {2014},
volume = {11},
number = {5},
pages = {877--884},
url = {https://mathopt.de/publications/Scholz2014.pdf}
}
@article{Engelhart2011,
author = {Engelhart, M. and Lebiedz, D. and Sager, S.},
title = {{O}ptimal {C}ontrol for {C}ancer {C}hemotherapy {ODE} {M}odels: {P}otential of optimal schedules and choice of objective function},
journal = {{M}athematical {B}iosciences},
year = {2011},
volume = {229},
number = {1},
pages = {123--134},
url = {https://mathopt.de/publications/Engelhart2011.pdf}
}
@mastersthesis{Engelhart2009,
author = {Engelhart, M.},
title = {{M}odeling, {S}imulation, and {O}ptimization of {C}ancer {C}hemotherapies},
school = {Heidelberg University},
year = {2009},
url = {https://mathopt.de/publications/Engelhart2009.pdf}
}
@article{Shaik2008,
author = {Shaik, O.S. and Sager, S. and Slaby, O. and Lebiedz, D.},
title = {{P}hase tracking and restoration of circadian rhythms by model-based optimal control},
journal = {{IET} {S}ystems {B}iology},
year = {2008},
volume = {2},
pages = {16--23}
}
@article{Koenig2006,
author = {K\"onig, R. and Schramm, G. and Oswald, M. and Seitz, H. and Sager, S. and Zapatka, M. and Reinelt, G. and Eils, R.},
title = {{D}iscovering functional gene expression patterns in the metabolic network of {E}scherichia coli with wavelets transforms},
journal = {{BMC} {B}ioinformatics},
year = {2006},
volume = {7},
pages = {119},
url = {http://www.biomedcentral.com/1471-2105/7/119}
}
@article{Lebiedz2005,
author = {Lebiedz, D. and Sager, S. and Bock, H.G. and Lebiedz, P.},
title = {{A}nnihilation of limit cycle oscillations by identification of critical phase resetting stimuli via mixed-integer optimal control methods},
journal = {{P}hysical {R}eview {L}etters},
year = {2005},
volume = {95},
pages = {108303}
}